Elana P. Simon

I’m excited about trying to understand what ML models are learning from biological sequences and structures - digging into their embeddings to find interpretable biological concepts, figuring out how they pick up these patterns during training, and seeing what biology we can uncover by reverse-engineering the models!
I’m doing a PhD at Stanford University, advised by James Zou. I also write ML-bio deep-dives on my blog (matmols), though these are pretty high-effort and thus low-frequency.
Previously I worked at Reverie Labs as an ML engineer helping design small molecule cancer drugs. As an undergrad, I studied Computer Science at Harvard and worked with Debora Marks on protein language models. I’ve also been quite involved with research, fundraising, and patient advocacy focused on Fibrolamellar Hepatocellular Carcinoma.
selected publications
-
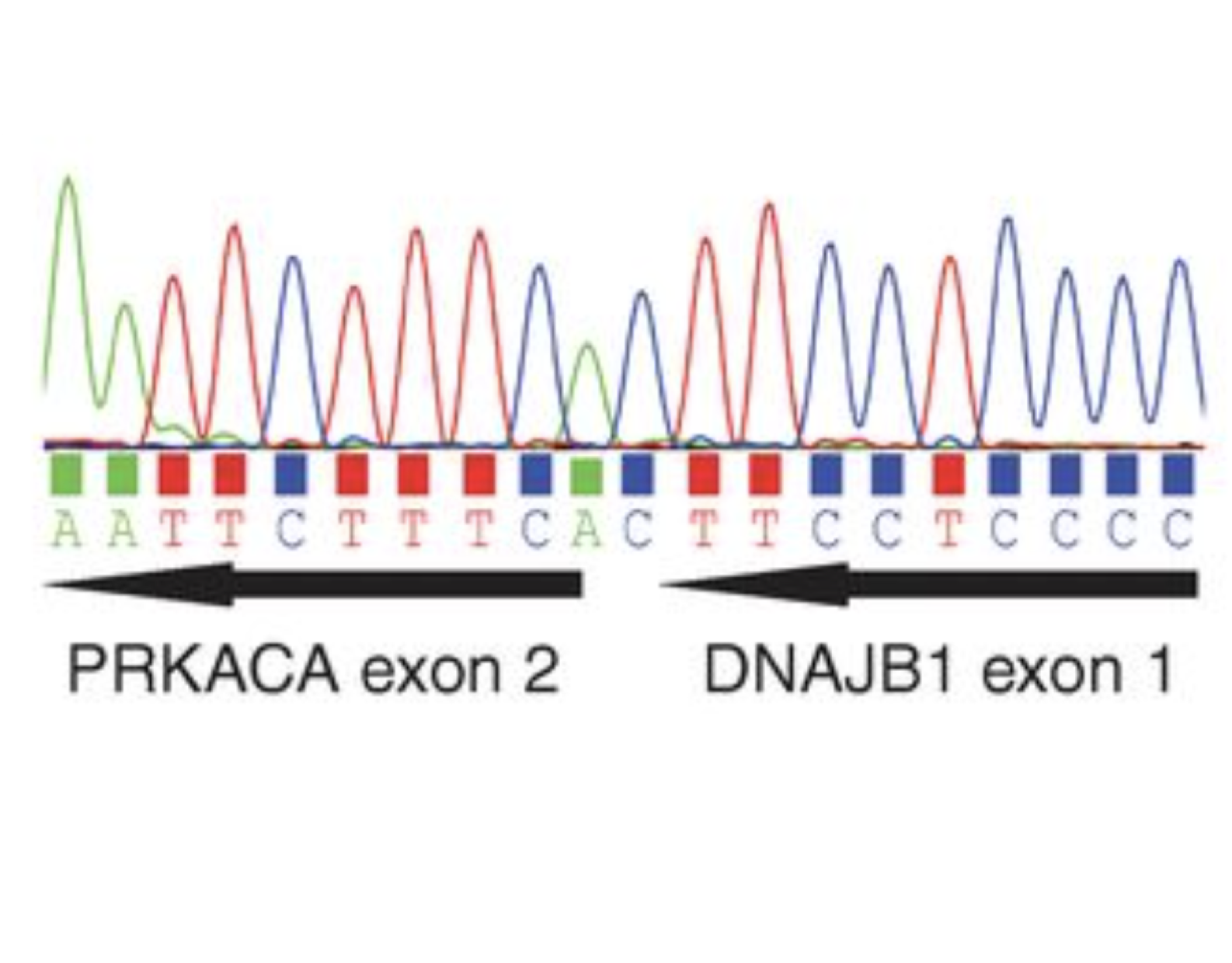 Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinomaScience, 2014
Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinomaScience, 2014 -
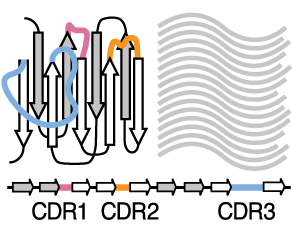 Protein design and variant prediction using autoregressive generative modelsNature communications, Apr 2021
Protein design and variant prediction using autoregressive generative modelsNature communications, Apr 2021 -
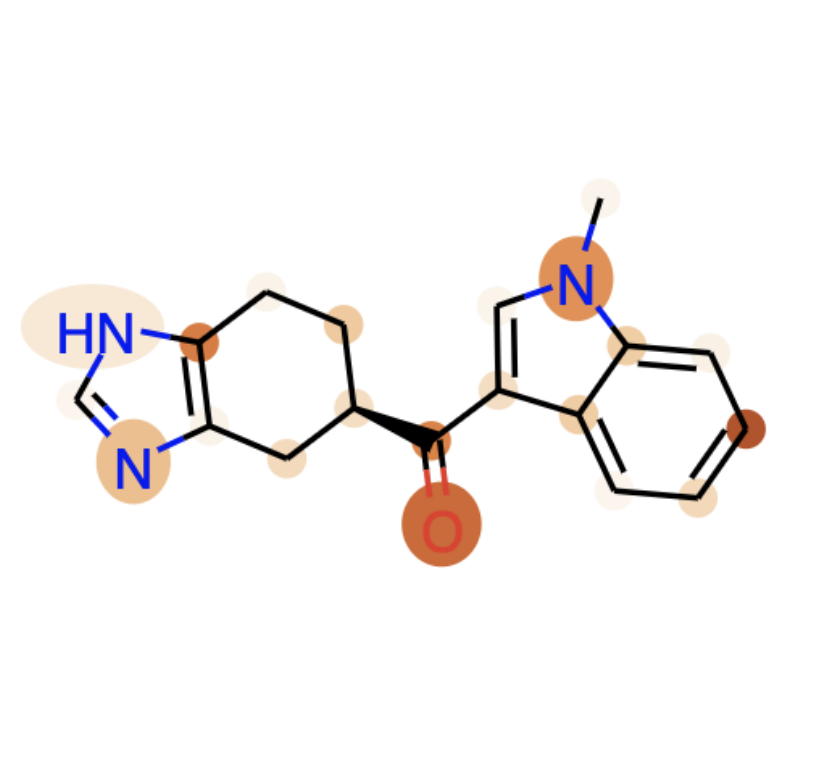 Chemberta-2: Towards chemical foundation modelsELLIS Machine Learning for Molecule Discovery Workshop, Dec 2021
Chemberta-2: Towards chemical foundation modelsELLIS Machine Learning for Molecule Discovery Workshop, Dec 2021 -
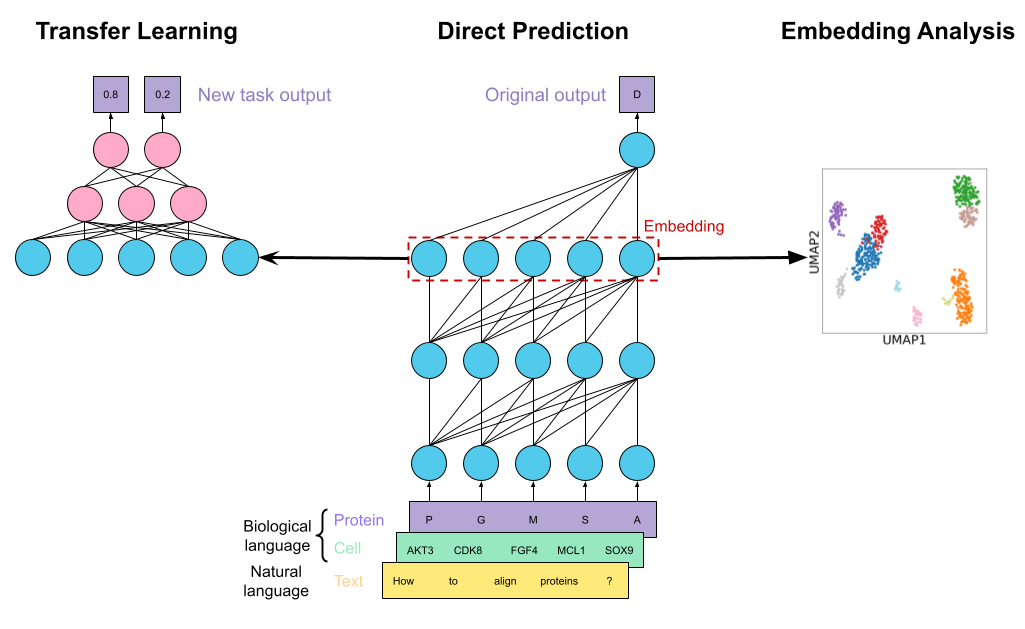 Language models for biological research: a primerNature Methods, Aug 2024
Language models for biological research: a primerNature Methods, Aug 2024 -
 InterPLM: Discovering Interpretable Features in Protein Language Models via Sparse AutoencodersbioRxiv, Nov 2024
InterPLM: Discovering Interpretable Features in Protein Language Models via Sparse AutoencodersbioRxiv, Nov 2024